#gene expression
Explore tagged Tumblr posts
Text
Ever since the COVID-19 pandemic kicked off, we've been far more aware of our sense of smell. Now, new research shows that odors – like those emanating from ripening fruits or fermented foods – can lead to changes in how genes are expressed inside cells far beyond the nose. The findings have scientists wondering if, with much more research, sniffing volatile, airborne compounds could be a way to treat cancer or slow neurodegenerative disease.
Continue Reading.
188 notes
·
View notes
Text

There is no context, and there will never be. The real context was the insulin we made along the way.
11 notes
·
View notes
Text
Entailed in Development
Hes gene activity, which oscillates, is crucial for differentiation of tissues in early embryo development. Here, Hes gene expression has been live-tracked in single cells of a developing mouse tail tip enabling its dynamics to be quantified for the first time
Read the published research article here
Video made with Leica Microsystems
Video from work by Yasmine el Azhar and colleagues
Hubrecht Institute-KNAW (Royal Netherlands Academy of Arts and Sciences), University Medical Center Utrecht, Utrecht, The Netherlands
Video originally published with a Creative Commons Attribution 4.0 International (CC BY 4.0)
Published in Development, September 2024
You can also follow BPoD on Instagram, Twitter and Facebook
#science#biomedicine#immunofluorescence#biology#embryo development#developmental biology#gene expression
8 notes
·
View notes
Text
Machine learning and the microscope
New Post has been published on https://thedigitalinsider.com/machine-learning-and-the-microscope/
Machine learning and the microscope


With recent advances in imaging, genomics and other technologies, the life sciences are awash in data. If a biologist is studying cells taken from the brain tissue of Alzheimer’s patients, for example, there could be any number of characteristics they want to investigate — a cell’s type, the genes it’s expressing, its location within the tissue, or more. However, while cells can now be probed experimentally using different kinds of measurements simultaneously, when it comes to analyzing the data, scientists usually can only work with one type of measurement at a time.
Working with “multimodal” data, as it’s called, requires new computational tools, which is where Xinyi Zhang comes in.
The fourth-year MIT PhD student is bridging machine learning and biology to understand fundamental biological principles, especially in areas where conventional methods have hit limitations. Working in the lab of MIT Professor Caroline Uhler in the Department of Electrical Engineering and Computer Science and the Institute for Data, Systems, and Society, and collaborating with researchers at the Eric and Wendy Schmidt Center at the Broad Institute and elsewhere, Zhang has led multiple efforts to build computational frameworks and principles for understanding the regulatory mechanisms of cells.
“All of these are small steps toward the end goal of trying to answer how cells work, how tissues and organs work, why they have disease, and why they can sometimes be cured and sometimes not,” Zhang says.
The activities Zhang pursues in her down time are no less ambitious. The list of hobbies she has taken up at the Institute include sailing, skiing, ice skating, rock climbing, performing with MIT’s Concert Choir, and flying single-engine planes. (She earned her pilot’s license in November 2022.)
“I guess I like to go to places I’ve never been and do things I haven’t done before,” she says with signature understatement.
Uhler, her advisor, says that Zhang’s quiet humility leads to a surprise “in every conversation.”
“Every time, you learn something like, ‘Okay, so now she’s learning to fly,’” Uhler says. “It’s just amazing. Anything she does, she does for the right reasons. She wants to be good at the things she cares about, which I think is really exciting.”
Zhang first became interested in biology as a high school student in Hangzhou, China. She liked that her teachers couldn’t answer her questions in biology class, which led her to see it as the “most interesting” topic to study.
Her interest in biology eventually turned into an interest in bioengineering. After her parents, who were middle school teachers, suggested studying in the United States, she majored in the latter alongside electrical engineering and computer science as an undergraduate at the University of California at Berkeley.
Zhang was ready to dive straight into MIT’s EECS PhD program after graduating in 2020, but the Covid-19 pandemic delayed her first year. Despite that, in December 2022, she, Uhler, and two other co-authors published a paper in Nature Communications.
The groundwork for the paper was laid by Xiao Wang, one of the co-authors. She had previously done work with the Broad Institute in developing a form of spatial cell analysis that combined multiple forms of cell imaging and gene expression for the same cell while also mapping out the cell’s place in the tissue sample it came from — something that had never been done before.
This innovation had many potential applications, including enabling new ways of tracking the progression of various diseases, but there was no way to analyze all the multimodal data the method produced. In came Zhang, who became interested in designing a computational method that could.
The team focused on chromatin staining as their imaging method of choice, which is relatively cheap but still reveals a great deal of information about cells. The next step was integrating the spatial analysis techniques developed by Wang, and to do that, Zhang began designing an autoencoder.
Autoencoders are a type of neural network that typically encodes and shrinks large amounts of high-dimensional data, then expand the transformed data back to its original size. In this case, Zhang’s autoencoder did the reverse, taking the input data and making it higher-dimensional. This allowed them to combine data from different animals and remove technical variations that were not due to meaningful biological differences.
In the paper, they used this technology, abbreviated as STACI, to identify how cells and tissues reveal the progression of Alzheimer’s disease when observed under a number of spatial and imaging techniques. The model can also be used to analyze any number of diseases, Zhang says.
Given unlimited time and resources, her dream would be to build a fully complete model of human life. Unfortunately, both time and resources are limited. Her ambition isn’t, however, and she says she wants to keep applying her skills to solve the “most challenging questions that we don’t have the tools to answer.”
She’s currently working on wrapping up a couple of projects, one focused on studying neurodegeneration by analyzing frontal cortex imaging and another on predicting protein images from protein sequences and chromatin imaging.
“There are still many unanswered questions,” she says. “I want to pick questions that are biologically meaningful, that help us understand things we didn’t know before.”
#2022#amazing#Analysis#Animals#applications#Autoencoders#bioengineering#Biology#Brain#Broad Institute#cell#Cells#China#chromatin#communications#computer#Computer Science#covid#data#deal#december#Disease#Diseases#engine#engineering#form#Forms#Fundamental#gene expression#genes
2 notes
·
View notes
Text
Diverse Headgear Of Hoofed Mammals Evolved From A Common Ancestor, study Baruch College & CUNY Graduate Center, published by Communications Biology
by @GrrlScientist
2 notes
·
View notes
Text
Multiple species of Tardigrades are resistant to drought, high doses of radiation, low oxygen environments, and both high and low temperatures and pressures.
Tardigrades transform into a dormant state called anhydrobiosis, which allows them to reversibly halt their metabolism.
This is a study of Tardigrades genes that allows this.
A large amount of research has focused on the genetic pathways related to these capabilities, and a number of genes have been identified and linked to the extremotolerant response of tardigrades
This study generate the first phylogenies of six separate protein families linked with desiccation and radiation tolerance in Tardigrada
2 notes
·
View notes
Text
Melanoma drug resistance get overcome: without GSK3 cells are B-Raft of advantage
Melanoma is a type of skin cancer in which nearly half of patients have mutations in the BRAF gene that accelerate tumor growth. Although BRAF-targeted treatments, known as BRAF inhibitors, work well initially, tumors often find ways to fight back. Adaptive metabolic reprogramming with oxidative phosphorylation, anaerobic glycolysis, and autophagy are the main mechanisms of resistance, both de…
#beta-catenin#cancer cells#dabrafenib#drug resistance#gene expression#melanoma#metastasis#protein kinase
0 notes
Text
Unlocking Insights: A Beginner’s Guide to Gene Expression Profiling and Pathway Enrichment
In the field of genomics, understanding the behavior of genes is essential to unlocking the mysteries of biology and disease. Two important techniques, gene expression profiling and pathway enrichment analysis, help researchers unravel the complexity of genetic regulation, allowing for a deeper understanding of biological processes, disease mechanisms, and therapeutic targets. This article provides a beginner’s guide to gene expression profiling and pathway enrichment analysis, explaining their significance, methods, and applications.
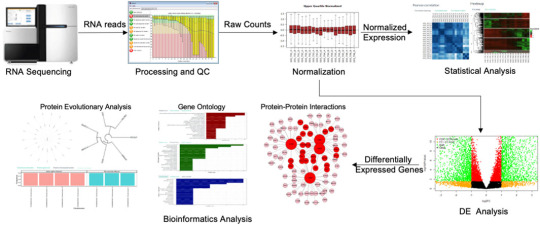
To explore more about applications of Gene Expression Profiling and Pathway Enrichment Analysis, you can visit Gene-omics.
If you want to learn about the genomics from the beginning, you can enroll for the Genome Analysis Bootcamp Demo Session here- https://edgeneomics.com/genome-analysis-bootcamp/
0 notes
Text
youtube
#Endometriosis#ferroptosis#FZD7#bioinformatics#gene expression#diagnostic biomarker#lipid peroxidation#oxidative stress#precision medicine#Wnt signaling#inflammation#reproductive health#molecular biology#transcriptomics#gene regulation#machine learning#pathway analysis#biomarker discovery#women’s health#gynecology.#Youtube
0 notes
Text
You Are Not Your Genes: How Lifestyle Choices Can Rewrite Your Biological Story
Imagine two identical twins, sharing the exact same DNA, yet one develops heart disease in their 50s while the other enjoys robust health well into their 80s. This scenario, while seemingly paradoxical, highlights the profound influence of epigenetics, a field revolutionizing our understanding of the interplay between genes and environment. Epigenetics, literally meaning “above genetics,”…
0 notes
Text
What are induced pluripotent stem cells, and how are they different from embryonic stem cells?
What are induced pluripotent stem cells (iPSCs)? Reprogrammed adult cells: iPSCs are created in the lab by taking adult cells (often skin or blood cells) and genetically reprogramming them back to an immature, embryo-like state. Pluripotency: Like embryonic stem cells, iPSCs are pluripotent. This means they have the exceptional potential to develop into almost any type of cell in the body. Key…

View On WordPress
#biotechnology#Cell Therapy#Cellular Differentiation#Cellular Reprogramming#CRISPR#Developmental Biology#Embryonic Stem Cells#ESCs#Ethical Considerations#gene editing#Gene Expression#Immune Response#Induced Pluripotent Stem Cells#iPSCs#Medical Research#Pluripotency#Regenerative medicine#Stem Cell Research
0 notes
Photo
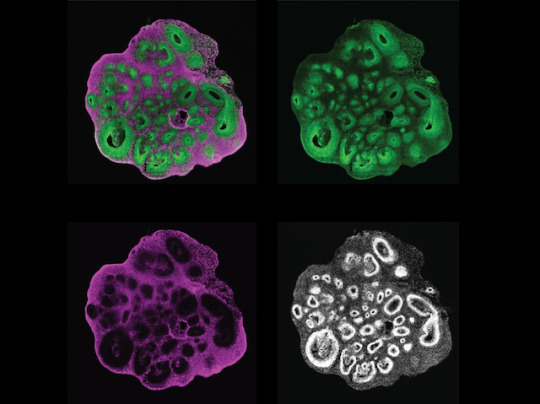
Exploring Family Trees
The cerebral cortex is the outermost layer of your brain, but creating this layered structure is a highly elaborate process during development. It requires early brain cell ancestors – called neural progenitor cells – to differentiate and create later generations of neurons and brain cells called glia. Integral to differentiation is ensuring that genes are switched on at the right time, but also that their genetic instructions are translated into an abundance of proteins to allow progenitors to become brain cells. The importance of these gene-protein networks and their timing during this process is known in mice, but remains unclear in humans. Researchers have now created a human brain organoid (pictured) that allows them to track progenitors (green) and neurons (magenta) simultaneously (all cell nuclei shown in white). This lab-grown ‘mini brain’ allows both cell types from different stages during development to be isolated and analysed, building a more detailed plan of this process.
Written by Sophie Arthur
Image from work by Jaydeep Sidhaye and Philipp Trepte, and colleagues
Institute of Molecular Biotechnology of the Austrian Academy of Sciences (IMBA), Vienna BioCenter (VBC), Vienna, Austria
Image originally published with a Creative Commons Attribution 4.0 International (CC BY 4.0)
Published in eLife, March 2023
You can also follow BPoD on Instagram, Twitter and Facebook
#science#organoids#biomedicine#neuroscience#neural progenitor cells#cells#immunofluorescence#brain#brain cells#neurons#gene expression#embryo development#developmental biology
14 notes
·
View notes
Text
Mouse Studies
First, it sniffed the unconscious mouse’s body and then started grooming the animal. Then it took more vigorous action: biting the mouth of its immobile partner and pulling its tongue out, clearing the airway opening.
The Mice had different squeak patterns than their generic partners. The mice used different sounds and higher frequency of squeaking noises. Tested in males wooing mates and infant mouse cries.
1 note
·
View note
Text
Born Again🐐🧬
By:
0 notes
Text
Bach 2 the skin: a-topic to explore to treat dermatitis making easier MAREs to trot
Atopic dermatitis is an allergy that affects an average of about 5% of the general population, with symptoms closely related to social stress. In socially active adults, the disease often becomes chronic. In the affected areas, infiltrating immune cells secrete inflammatory cytokines, which contribute to the development of symptoms. While these cytokines normally play a protective role against…
#atopic dermatitis#gene expression#immune response#inflammation#interleukin-2#protein kinase#signaling pathway#skin barrier#T cells#transcription factors
0 notes
Text
Real-World Applications of Gene Expression Profiling and Pathway Enrichment in Cancer Research
Cancer remains one of the leading causes of death worldwide, prompting ongoing research to uncover its underlying mechanisms and develop effective treatments. Among the numerous tools available to cancer researchers, gene expression profiling and pathway enrichment analysis have emerged as powerful methods for understanding tumor biology, identifying therapeutic targets, and improving patient outcomes. This article explores the real-world applications of gene expression profiling in cancer research, highlighting how these techniques are reshaping the landscape of cancer diagnosis and treatment.

To explore more about applications of Gene Expression Profiling and Pathway Enrichment Analysis you can visit Gene-omics.
If you want to learn about the genomics from the beginning, you can enroll for the Genome Analysis Bootcamp Demo Session here- https://edgeneomics.com/genome-analysis-bootcamp/
To learn Gene Expression Analysis, you may visit https://edgeneomics.com/gene-expression-with-r-masterclass/
0 notes